The stabilization, remodelling, and repair of biological membranes are critical tasks by which eukaryotic cells maintain cellular compartmentalization, cope with membrane damage, and ensure controlled transmembrane transport of molecules and ions. In eukaryotes, several membrane remodelling processes involve the ESCRT-III protein, the core component of the endosomal sorting complexes required for transport (ESCRT). Recently, ESCRT-III proteins have been shown to be also conserved in bacteria, and the ESCRT-III superfamily member IM30 (inner membrane-associated protein of 30 kDa) binds to, stabilizes and remodels membranes. These observations now enable us to study the bacterial system in detail, to delineate conserved molecular mechanisms of ESCRT-III-mediated membrane remodelling.
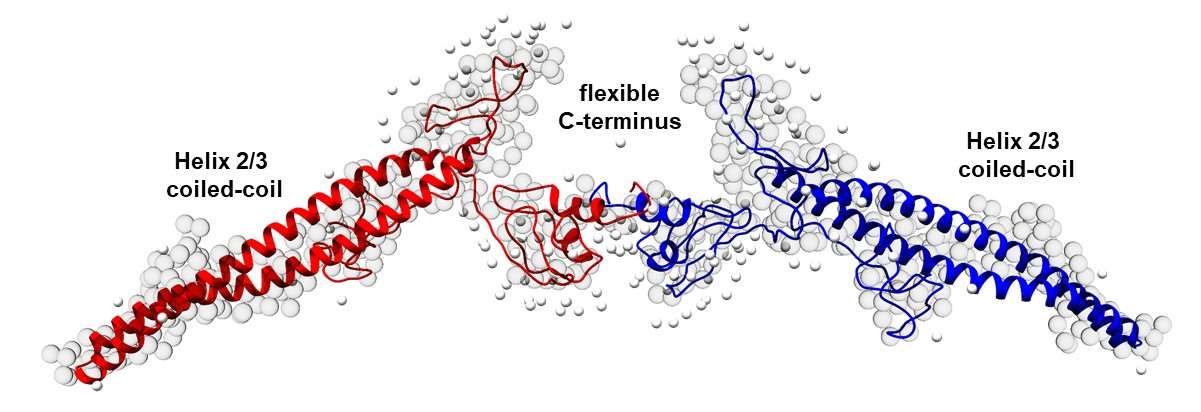
IM30 monomers assemble to form large homo-oligomeric barrel structures, in which the monomers are ~80% α‑helical. Upon membrane binding, the barrels disassemble, and the C-terminal region of monomeric IM30 unfolds. Thus, inter-monomer contacts in the barrel apparently induce the formation of α-helices. Furthermore, upon barrel disassembly, IM30 forms a membrane-covering “carpet” that stabilizes the membrane. In solution, oligomerization-impaired IM30 and the disordered IM30 C terminus alone form liquid condensates by a process of liquid–liquid phase separation. Thus, the formation of IM30 barrels not only induces α-helix formation but also appears to prevent liquid–liquid phase separation. We anticipate that increasing the concentration of unfolded IM30 on a membrane surface above the saturating concentration required for phase separation promotes phase separation and formation of a 2D condensate on the membrane surface; this is reminiscent of the large assemblies observed in vivo on membrane surfaces. In this project, we will study the α‑helical-to-disordered structural transition of IM30 in solution and on membranes surfaces, using a combination of molecular dynamics simulations, biochemistry and biophysical analyses involving advanced spectroscopic methods. We will link primary sequence features and the physical characteristics of condensates with the putative physiological IM30 functions, using a sequence-directed computational and experimental analysis.